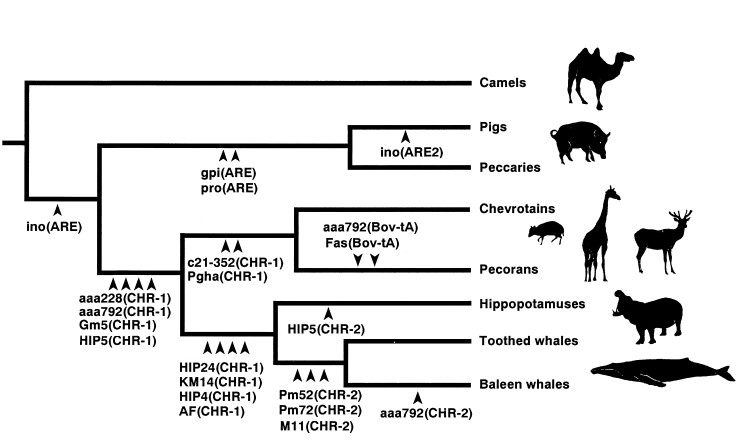
The diagram below shows a phylogeny of artiodactyls ("even toed" mammals), as deduced from 20 different SINEs (Short INterspersed Elements) and LINEs (Long INterspersed Elements) in their junk DNA. (To understand this, you might want to read my short summary of the theory.)
Each arrowhead means that all of the species to its right have one particular SINE or LINE at one particular locus (place in the genome). For example, the arrow marked "ino(ARE2)" means that the "ARE2" pattern was found at the "ino" locus, but only in pigs. The arrows marked "gpi(ARE)" and "pro(ARE)" mean that the "ARE" pattern was found at two places in the Pig genome, and at the exact same two places in the Peccary genome.
Notice that this phylogeny is the only way to connect these species without assuming huge coincidences. And actually, the data is stronger than this diagram shows. The authors tested four Pecorans - Sheep, Cow, Reticulated Giraffe, and Axis Deer.
The diagram is from:
Phylogenetic relationships among cetartiodactyls based on insertions of short and long interspersed elements: Hippopotamuses are the closest extant relatives of whales, Nikaido, Rooney and Okada, Proceedings of the National Academy of Sciences 96, 10261 (31 August 1999)